The GLUE study (Multi-omics single-cell data integration and regulatory inference with graph-linked embedding) published in Nature Biotechnology was recently awarded by Genomics, Proteomics and Bioinformatics (GPB) as China’s top ten bioinformatics advances of 2022.
With rapid advances in recent years, single-cell omics technologies have become essential tools for studying cellular function and gene regulation. Up until now, many omics layers can be profiled at single-cell resolution, each of which reflects one specific aspect of the cellular state. To achieve more holistic characterization of cellular states as well as the underlying regulatory circuits, Gao lab developed a novel computational model called GLUE. The GLUE model employs the variational autoencoder (VAE) to learn low-dimensional cell embeddings. Due to the discrepancy in feature spaces, different autoencoders are required for different omics layers. In order to link the omics-specific autoencoders, a graph-linking strategy is proposed. Specifically, prior knowledge regarding regulatory interactions between omics features is encoded into a guidance graph, effectively linking different autoencoders to ensure “semantic consistency” of the learned cell embeddings (Fig. 1). GLUE achieves high integration accuracy, robustness and scalability, supports the integration of arbitrary numbers of omics layers with arbitrary regulatory directions, and is capable to integrative regulatory inference at the same time.
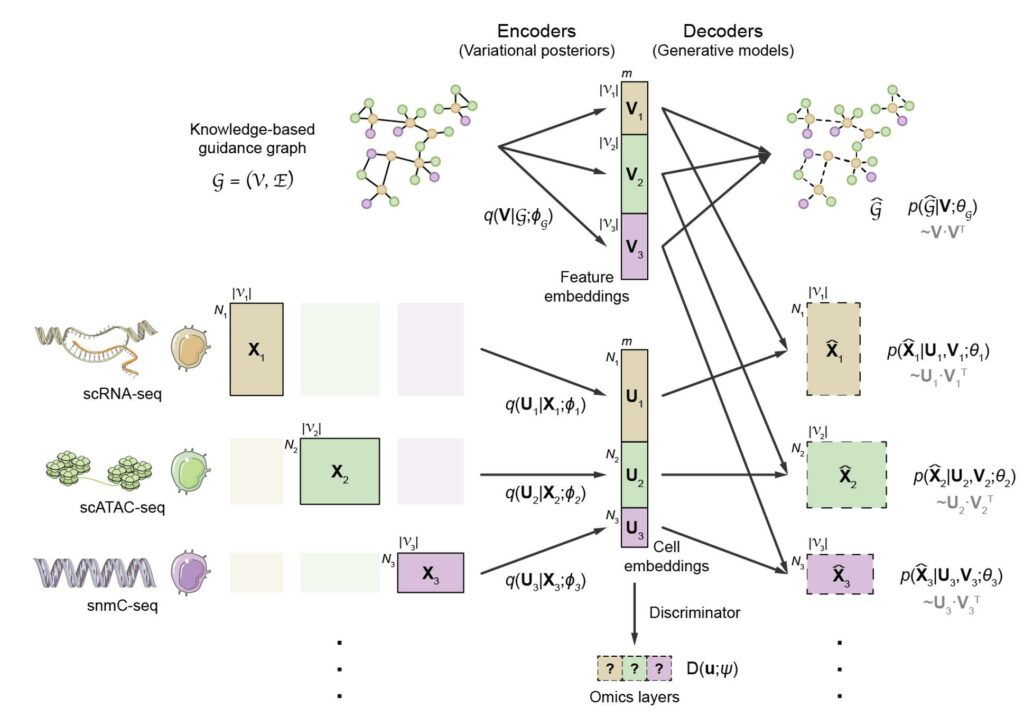
Fig. 1 Architecture of the GLUE model.
The “China’s top ten bioinformatics” series was initiated by Genomics, Proteomics and Bioinformatics (GPB) from 2018, aimed at promoting innovation and showcasing major advances in China’s bioinformatics research. Previously, the human lncRNA atlas and singe-cell transcriptomics cell querying studies from Gao lab was awarded as China’s top ten bioinformatics database of 2019, and top ten bioinformatics advances of 2020, respectively.
Application URL:
https://github.com/gao-lab/GLUE
Publication:
Cao, Z.-J. & Gao, G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 2022;40:1458-66. PMID: 35501393