Through the process of transcription and translation, protein-coding genes (hereinafter referred to as genes) can guide the synthesis of proteins that are crucial to life activities, and then affect the physiological or pathological traits of living organisms. Thus, genes are the basic functional units in cells. Research in the past decades has shown that in the process of evolution, organisms can acquire new genes to perform new functions. As a matter of fact, new genes birth is one of the important driving forces that mediate the phenotypic evolution of species.
All things are born and die. In contrast to the emerging new genes, existing protein-coding genes may also lose their original functions due to mutation events such as insertions or deletions, and then be lost from the genome. However, limited by many factors, there has been a lack of systematic research on gene loss and its impact for a long time.
Recently, Gao Lab from Biomedical Pioneering Innovation Center (BIOPIC), Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI), and State Key Laboratory of Protein and Plant Gene Research at School of Life Sciences, developed a novel gene loss identification pipeline LOST & FOUD (LOcal Sequence-based Tracing Functional Ortholog UNit Death) and systematically annotated gene loss events in humans based on this pipeline. This work has been published in Molecular Biology and Evolution.

In order to facilitate the systematic identification of gene loss, Gao Lab proposed a novel gene loss identification pipeline that integrates orthologous inference in multiple species and genome alignment, LOST & FOUND (Figure 1). With the usage of genome alignment, LOST & FOUND is more capable of identifying the loss events suffering from large-scale deletion while the usage of orthologous inference in multiple species enables LOST & FOUND to effectively distinguish gene gain in reference species from gene loss in query species.
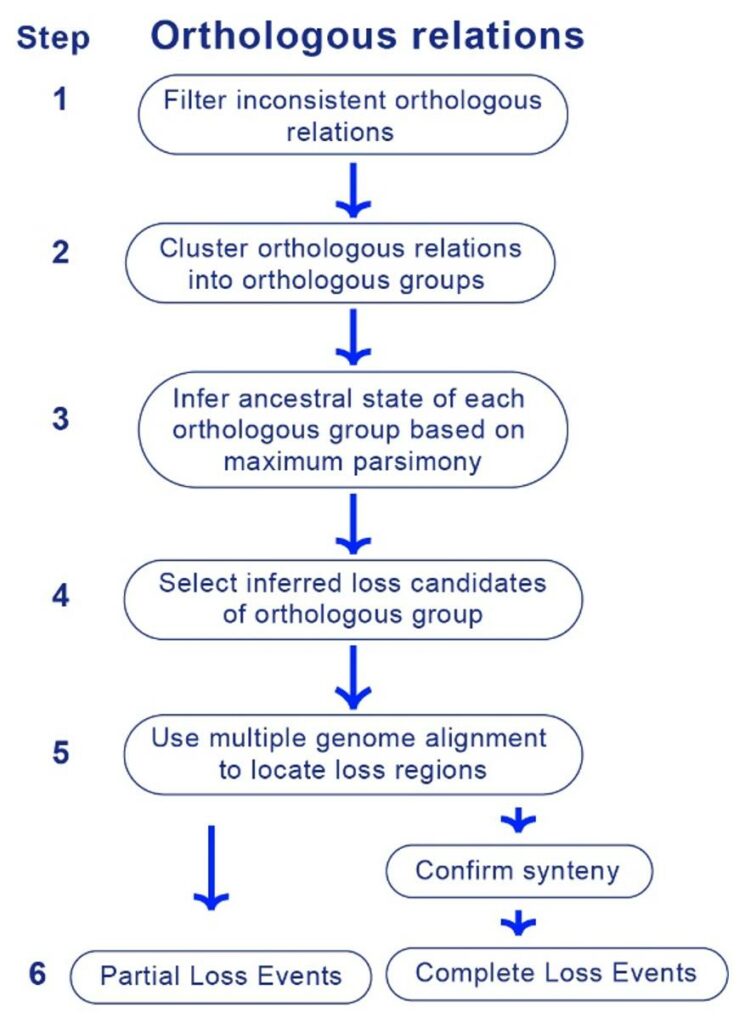
Figure 1. Workflow of LOST & FOUND.
Based on this pipeline, the team identified 155 gene loss events in humans, 88 of which contained loss relics in the human genome. Interestingly, by comparing these relics with annotated lncRNAs, the team found that 33 gene loss events were associated with the origin of lncRNAs (Figure 2 A-B), which were named Derived lncRNAs.
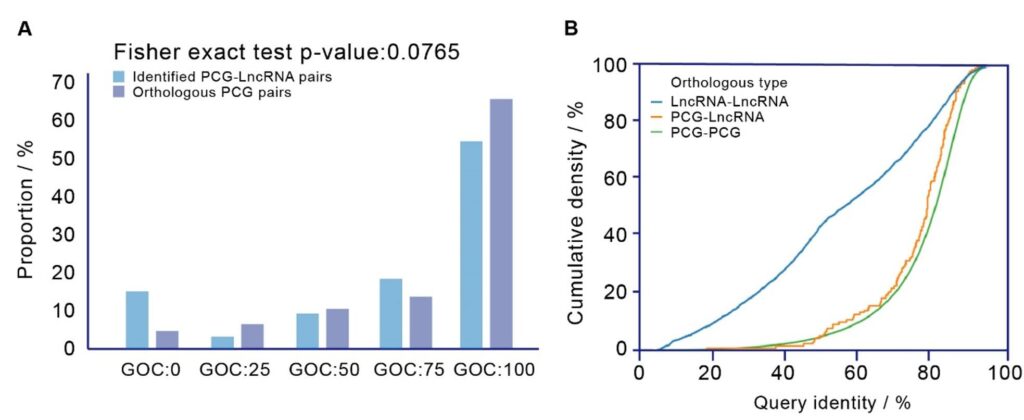
Figure 2. Orthologous relations between derived lncRNAs and gene loss events.
Derived lncRNAs are different from other lncRNAs, in that Derived lncRNAs are highly and broadly expressed, significantly longer, and more conserved (Figure 3 A-F). Besides, functional analyses suggested that these Derived lncRNAs are involved in growth, development, immunity, reproduction, and tumor-suppressive processes. In addition, over half (17 out of 33) of these Derived lncRNAs are under positive selection.
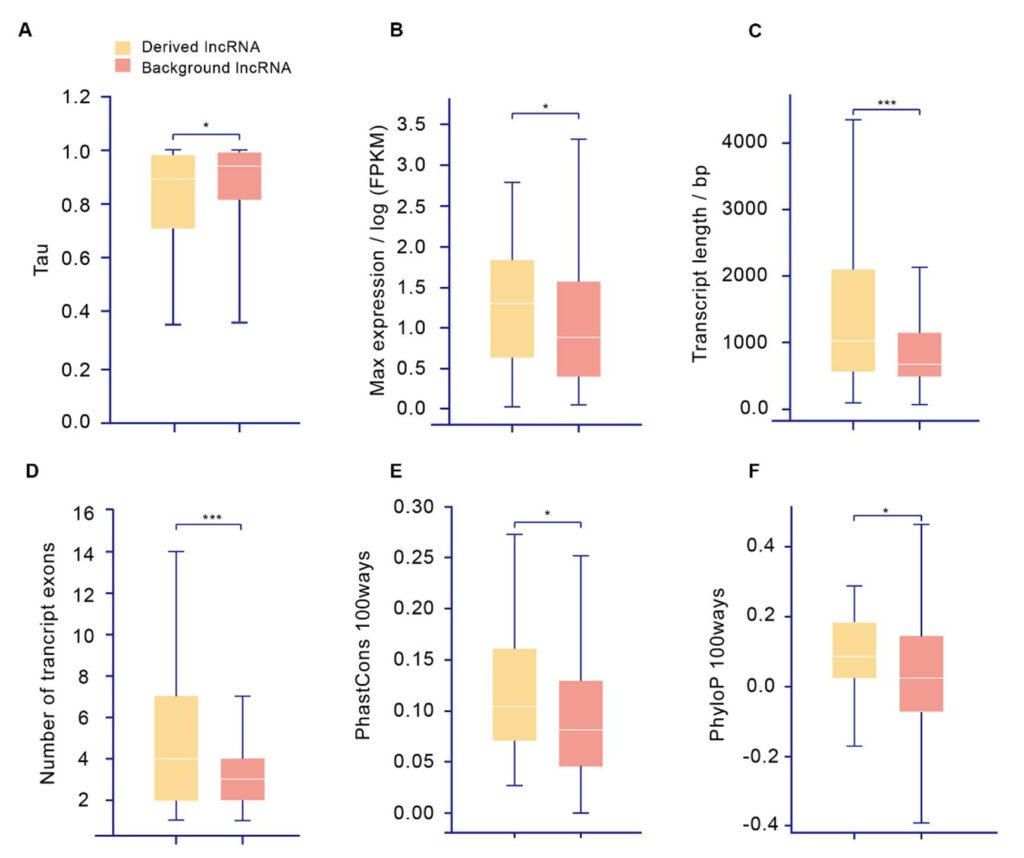
Figure 3. Comparison between Derived lncRNAs and other lncRNAs.
In summary, Gao Lab developed a new pipeline for gene loss identification and systematically annotated gene loss events in humans. It is worth noting that 33 gene loss events were related to the origin of lncRNAs, and these lncRNAs may carry important functions. Combined with the previous work [1-3], these results suggest that the loss of protein-coding genes could undergo “rebirth” as functional lncRNAs, which brings interesting connections between gene life and death or between coding and non-coding.
PhD student Zheng-Yang Wen is the first author of the study. Professor Ge Gao is the corresponding author. The study was supported by funds from the National Key Research and Development Program, the State Key Laboratory of Protein and Plant Gene Research, the Beijing Advanced Innovation Center for Genomics at Peking University. Part of the analysis was carried out on the Computing Platform of the Center for Life Sciences of Peking University and supported by the High-performance Computing Platform of Peking University.
Link:https://academic.oup.com/mbe/advance-article/doi/10.1093/molbev/msad103/7151558?login=true
References:
1.Duret, L., Chureau, C., Samain, S., Weissenbach, J. & Avner, P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312, 1653-1655 (2006). https://doi.org:10.1126/science.1126316
2.Zhao, Y. et al. Identification and analysis of unitary loss of long-established protein-coding genes in Poaceae shows evidences for biased gene loss and putatively functional transcription of relics. BMC Evol Biol 15, 66 (2015). https://doi.org:10.1186/s12862-015-0345-x
3.Hezroni, H. et al. A subset of conserved mammalian long non-coding RNAs are fossils of ancestral protein-coding genes. Genome Biol. 18, 162 (2017). https://doi.org:10.1186/s13059-017-1293-0