Long noncoding RNAs (lncRNAs) have been demonstrated to play many important regulatory roles via various mechanisms (1), and many characteristics have been used to infer their functions. For example, analyzing the natural selection pressures on lncRNAs can reveal whether they are functional (2), and their subcellular localization indicates where they perform functions (3). Functional enrichment of protein-coding genes whose expression profiles are highly correlated with lncRNAs provides important hints for potential lncRNA functionality (4). In addition, it has been reported that many lncRNAs perform their functions by interacting with other cellular factors; thus, studying interacting partners of lncRNAs, e.g. proteins and miRNAs, provides insights into their possible molecular mechanisms (5, 6). Though several databases are available for known lncRNAs (7, 8), with the abundant mammalian lncRNAs identified recently, a comprehensive annotation resource for these novel lncRNAs is still an urgent need.
Since its first release in November 2016, AnnoLnc (9) has been the only online server for comprehensively annotating novel human lncRNAs on-the-fly. Here, with significant updates to multiple annotation modules, backend datasets and the code base, AnnoLnc2 continues the effort to provide the scientific community with a one-stop online portal for systematically annotating novel human and mouse lncRNAs with a comprehensive functional spectrum, covering sequence and structure, expression and regulation, function and interaction, as well as genetic association and evolution (Figure 1). We believe that updated AnnoLnc2 (http://annolnc.gao-lab.org/) will help both computational and bench biologists identify lncRNA functions and investigate underlying mechanisms.
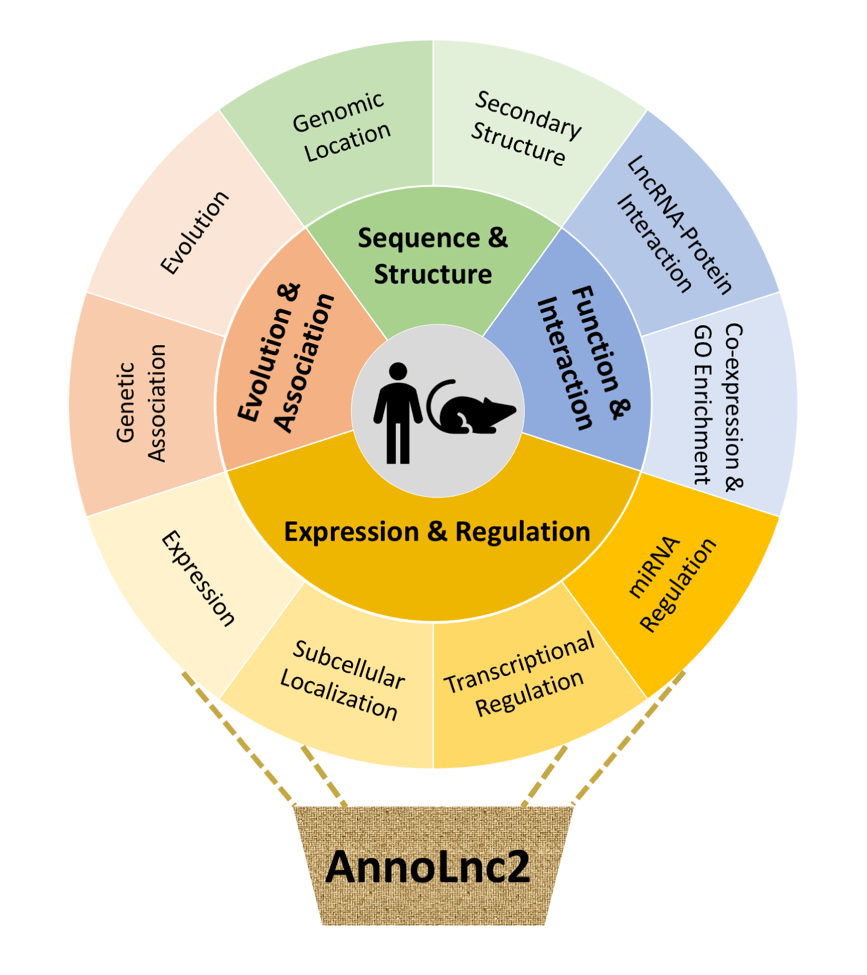
Figure 1. Framework of AnnoLnc2.
As the successor of AnnoLnc, AnnoLnc2 supports annotating both human and mouse novel lncRNAs with wider and more comprehensive data resources. The expression module supports simultaneous expression calling in 95 human samples (covering 26 normal tissues, 39 cancer cell lines and 2 normal cell lines) or 69 mouse samples (covering 28 normal tissues and 7 normal cell lines) for any lncRNA in nearly real time. The transcriptional regulation module integrates millions of ChIP-seq-based binding sites for 1339 human transcription factors (TFs) and 738 mouse TFs. By integrating 385 human CLIP-seq datasets (for 188 RNA binding proteins, RBPs) and 238 mouse CLIP-seq datasets (for 62 RBPs), the protein interaction module provides the most comprehensive online resource for CLIP-seq-based lncRNA–protein interaction annotation so far. Please also see more detailed annotation resources at http://annolnc.gao-lab.org/help.php#link-update.
AnnoLnc2 takes RNA sequences in fasta format as input, and users can paste or upload sequences on the AnnoLnc2 homepage and choose corresponding species from a drop-down option list. After that, users simply need to click the ‘GO’ button, and AnnoLnc2 will analyze all inputted sequences (up to 100 sequences) simultaneously (Figure 2A). After a job is submitted, AnnoLnc2 will return a link to users for retrieving results in the future. AnnoLnc2 provides an item for each best alignment of all sequences, and users can view detailed annotation results from the link in the ‘Status’ column. All of the results are shown in interactive figures and tables and allow users to view detailed numeric information and search for items of interest, such as TFs, RBPs, traits, or functions. To make using AnnoLnc2 more convenient, AnnoLnc2 supports batch uploading and downloading to avoid laboring operations for fetching results one by one (Figure 2B). Moreover, AnnoLnc2 also provides a standalone version for large-scale offline analysis (available from http://annolnc.gao-lab.org/download.php).
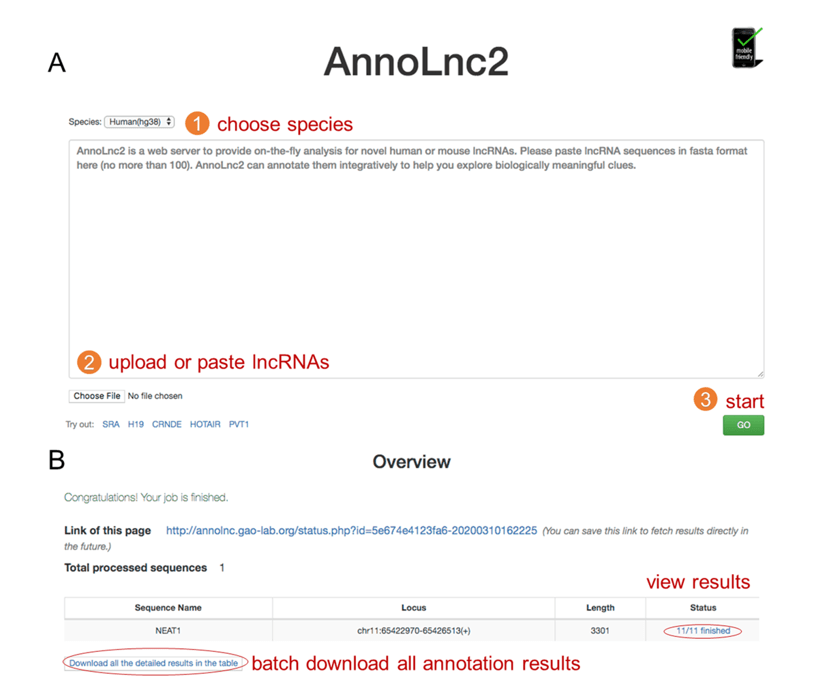
Figure 2. AnnoLnc2 web server. Users can run AnnoLnc2 through a three-step operation (A) and view detailed annotation results, as well as download all results by one click (B).
This research was published online in Nucleic Acids Research on May 14, 2020. Lan Ke and De-Chang Yang, Ph.D. students of the School of Life Sciences of Peking University, are the co-first authors of this paper. Dr. Ge Gao, a researcher at the School of Life Sciences of Peking University is corresponding authors. The research was supported by National Key Research and Development Program; China 863 Program; State Key Laboratory of Protein and Plant Gene Research and the Beijing Advanced Innovation Center for Genomics (ICG) at Peking University; National Program for Support of Top-notch Young Professionals (to G.G.).
Web site: http://annolnc.gao-lab.org/
Paper link: https://doi.org/10.1093/nar/gkaa368
References:
- Marchese,F.P., Raimondi,I. and Huarte,M. (2017) The multidimensional mechanisms of long noncoding RNA function. Genome Biol., 18, 206.
- Ponjavic,J., Ponting,C.P. and Lunter,G. (2007) Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res., 17, 556–65.
- Zhang,K., Shi,Z.M., Chang,Y.N., Hu,Z.M., Qi,H.X. and Hong,W. (2014) The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene, 547, 1–9.
- Guttman,M., Amit,I., Garber,M., French,C., Lin,M.F., Feldser,D., Huarte,M., Zuk,O., Carey,B.W., Cassady,J.P., et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature, 458, 223–7.
- Yoon,J.-H., Abdelmohsen,K. and Gorospe,M. (2014) Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol., 34, 9–14.
- Castello,A., Fischer,B., Eichelbaum,K., Horos,R., Beckmann,B.M., Strein,C., Davey,N.E., Humphreys,D.T., Preiss,T., Steinmetz,L.M., et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell, 149, 1393–406.
- Ma,L., Cao,J., Liu,L., Du,Q., Li,Z., Zou,D., Bajic,V.B. and Zhang,Z. (2019) LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res., 47, 2699.
- Fang,S., Zhang,L., Guo,J., Niu,Y., Wu,Y., Li,H., Zhao,L., Li,X., Teng,X., Sun,X., et al. (2018) NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res., 46, D308–D314.
- Hou,M., Tang,X., Tian,F., Shi,F., Liu,F. and Gao,G. (2016) AnnoLnc: a web server for systematically annotating novel human lncRNAs. BMC Genomics, 17, 931.