Long noncoding RNAs (lncRNAs) are defined as non-coding transcripts longer than 200 nt (1,2). They have been demonstrated to conduct diverse functions in multiple biological and pathological processes (3). A high-quality and comprehensive lncRNA annotation is a cornerstone requirement of subsequent functional investigation. However, large discrepancies still exist in the current major annotations.
By analyzing the largest compendium of 14,166 samples from normal and tumor tissues, we significantly expand the landscape of human long noncoding RNA with a high-quality atlas: RefLnc (Reference catalog of LncRNA, http://reflnc.gao-lab.org/). In addition to verifying 50,380 known lncRNAs, we have identified 27,520 novel lncRNA transcripts in which 83.6% are intergenic (Figure 1). And 91.4% (52 multi-exon and 33 single-exon) are successfully validated by Sanger sequencing independently out of 93 selected novel intergenic lncRNAs.
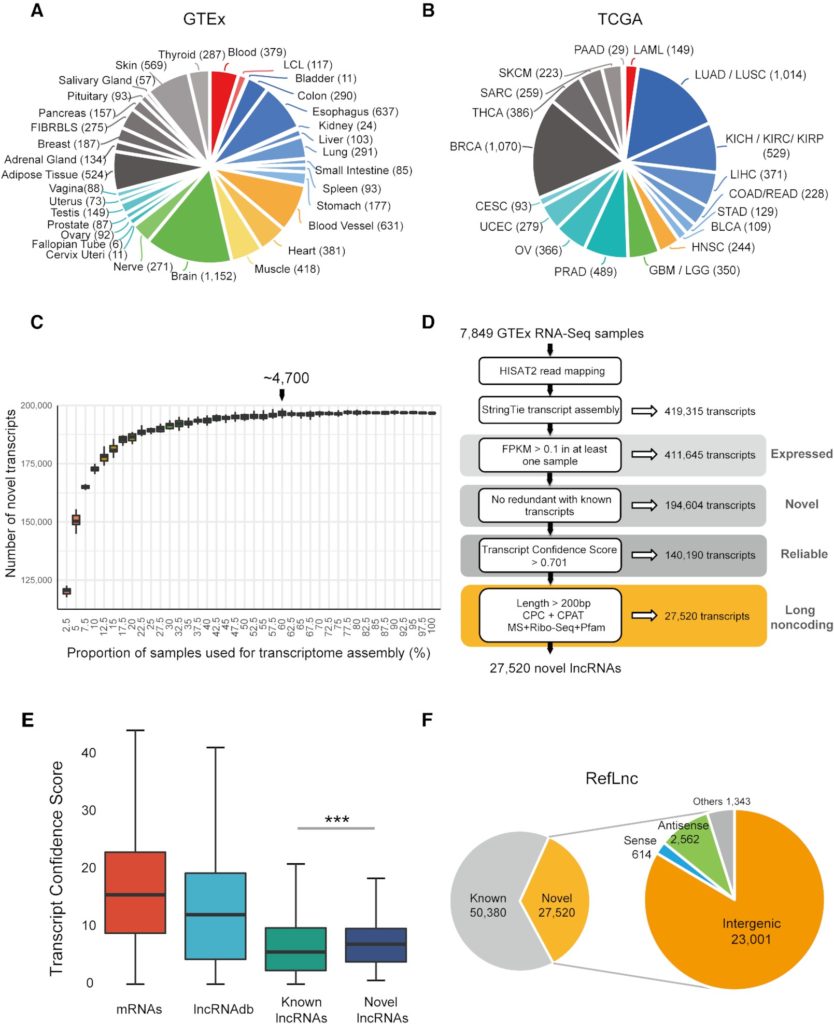
Figure 1. Reference-guided transcriptome assembly greatly expands the landscape of human lncRNAs. (A) The composition of the 7,849 physiological samples of 30 physiological tissues and two cell lines used for transcriptome reconstruction. (B) The composition of the 6,317 samples of 18 tumors from TCGA. (C) The number of novel transcripts assembled using different size of sample sets. Each dataset includes all type of tissues, sexes and races. (D) An integrative computational pipeline for lncRNA identification. (E) The Transcript Confidence Score (TCS) of novel lncRNAs is higher than that of known lncRNAs. (F) In total, RefLnc contains 77,900 lncRNAs including the verified known and novel lncRNAs, and 83.6% of the novel lncRNAs are in intergenic regions.
Consistent with previous reports, lncRNAs have lower expression and less alternative splicing efficiency than protein-coding genes, and lncRNAs are expressed in a much more tissue-specific manner than mRNAs (Figure 2). Moreover, we detect 75 novel lincRNAs with strongly sex-biased expression patterns, 132 novel lincRNAs whose expression levels are globally associated with age and 70 novel lincRNAs differentially expressed among individuals of various races. Last but not least, 160 novel lincRNAs are overlapped with 189 intergenic SNPs reported in 159 genome-wide association studies.
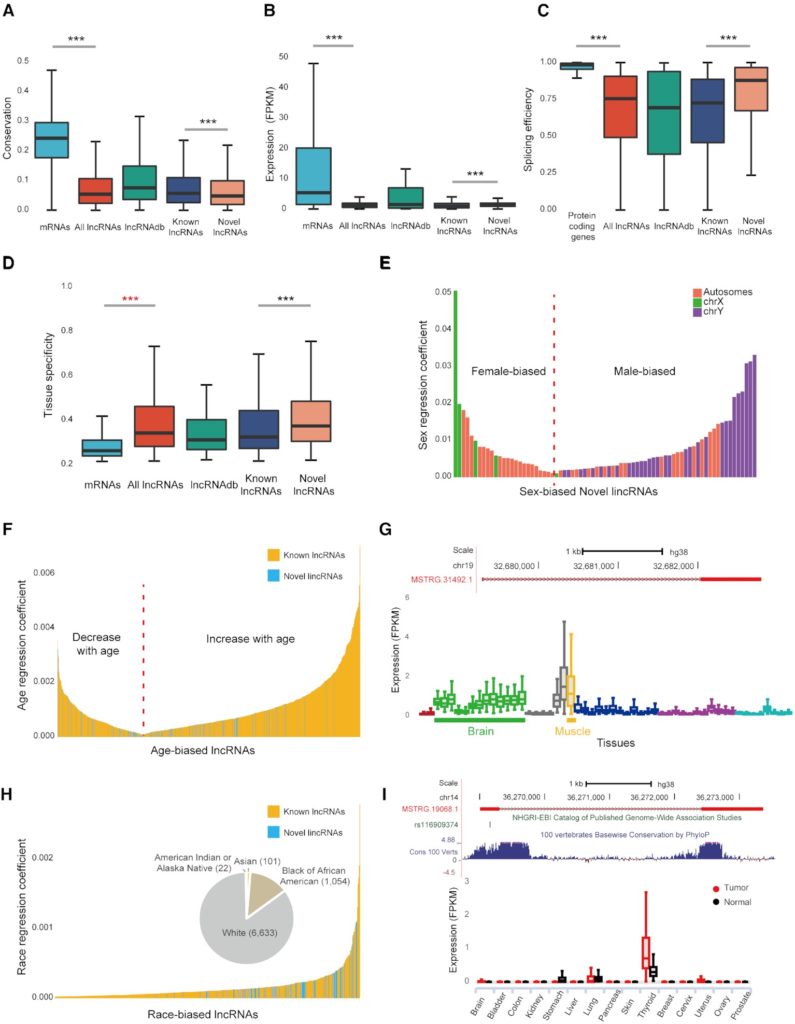
Figure 2. Characterization of the RefLnc assembly. (A) The conservation of lncRNAs is lower than that of mRNAs. (B) The expression levels of lncRNAs are lower than that of mRNAs. (C) lncRNAs have lower splicing efficiency than protein-coding genes. (D) lncRNAs are expressed in a much more tissue-specific manner than mRNAs. (E) Sex-biased novel lincRNAs that are differentially expressed between males and females (FDR < 0.05). (F) Novel lincRNAs and known lncRNAs correlated with age (FDR < 0.001). (G) The genomic view and expression patterns in normal samples of the age-associated novel lincRNA MSTRG.31492.1. (H) Novel lincRNAs and known lncRNAs that are differentially expressed across different races (FDR < 0.05). (I) The genomic view and differential expression patterns between tumors and normal tissues of the novel lincRNA MSTRG.19068.1, which overlaps a thyroid cancer risk-associated SNP.
To further explore the potential roles of our newly detected lncRNAs in cancer development, we scan 6,317 tumor samples across 18 tumors in TCGA and investigate novel lincRNAs associated with clinical outcomes such as tumor metastasis, recurrence, clinical stage and survival (Figure 3). For example, 180 novel lincRNAs are associated with overall survival time in the brain tumor and one-half (47.2%, 76/161) are expressed and validated in the independent Chinese LGG dataset of 258 glioma samples.
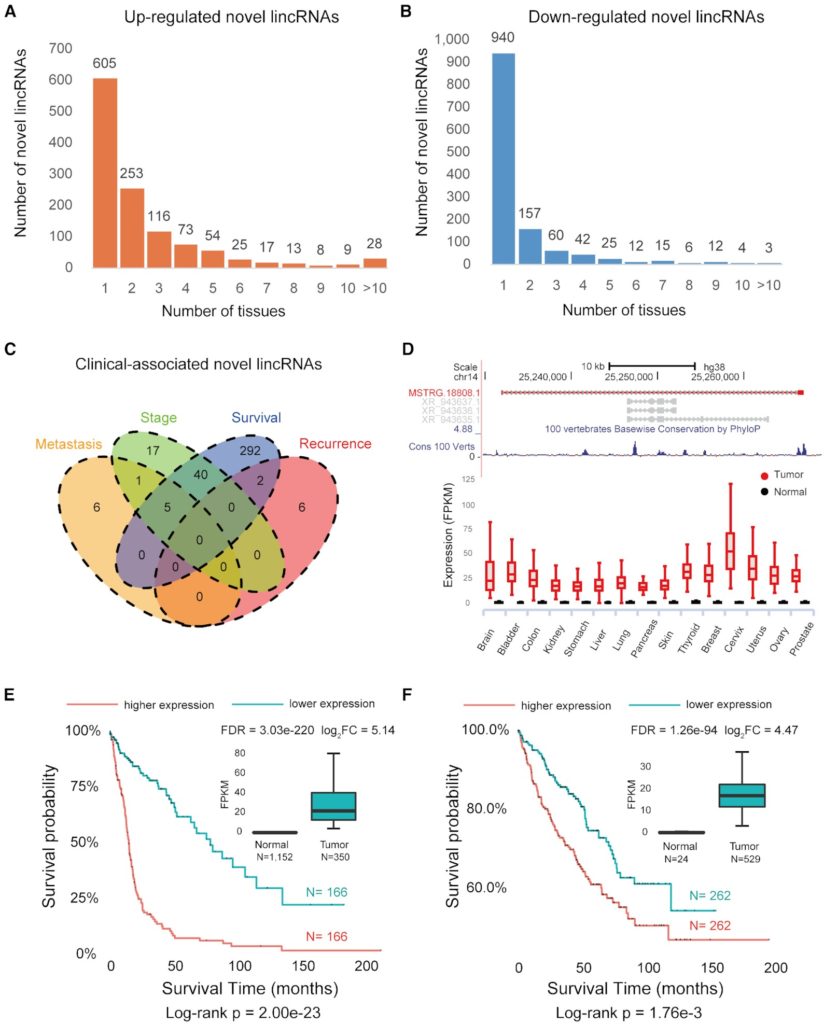
Figure 3. Discovery of tumor-associated novel lincRNAs. (A) Novel lincRNAs that are up-regulated in various tumors. (B) Novel lincRNAs that are down-regulated in various tumors. (C) The Venn diagram of clinical-associated novel lincRNAs. (D) The genomic view and differential expression pattern of the survival-associated novel lincRNA MSTRG.18808.1. (E) The expression of MSTRG.18808.1 is associated with poorer patient survival in the brain tumor. (F) The expression of MSTRG.18808.1 is correlated with poorer patient survival in the kidney tumor.
To facilitate the usage of RefLnc by the wider research community, we develop an online portal for visualizing the detailed characteristics of lncRNAs in 7,849 normal samples and 6,317 tumor samples (Figure 4).
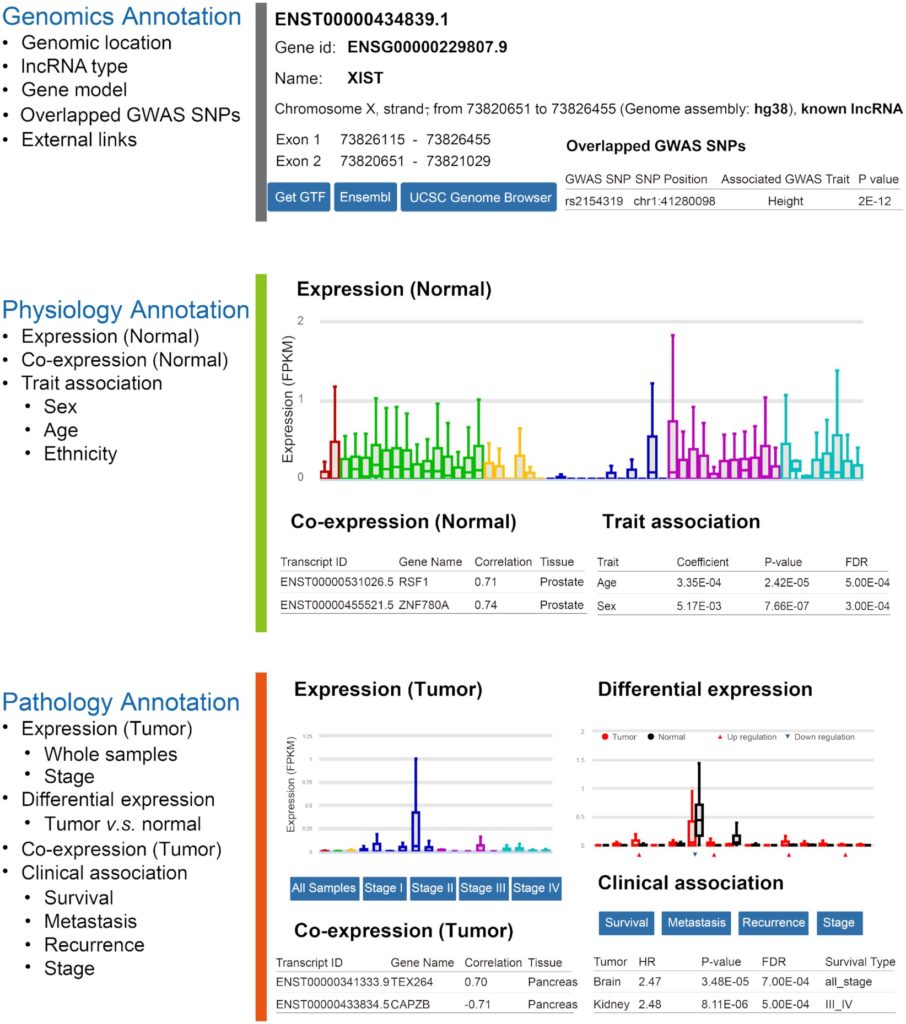
Figure 4. The architecture of the online webserver RefLnc. It provides detailed annotation of each lncRNA in RefLnc including genomics annotation, physiology annotation and pathology annotation.
Overall, RefLnc has greatly expanded the landscape of human lncRNAs and enabled the genome-wide exploration of the physiological function and clinical significance of lncRNAs. We anticipate that the RefLnc assembly as well as the computational pipelines developed will help to advance our knowledge of lncRNAs and provide a foundation for lncRNA genomics and biomarker development.
Website: http://reflnc.gao-lab.org/
Paper link: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkz621/5539882#138420864
References:
- Kapranov, P., Cheng, J., Dike, S., Nix, D.A., Duttagupta, R., Willingham, A.T., Stadler, P.F., Hertel, J., Hackermuller, J., Hofacker, I.L. et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science, 316, 1484-1488.
- Mattick, J.S. and Rinn, J.L. (2015) Discovery and annotation of long noncoding RNAs. Nature structural & molecular biology, 22, 5-7.
- Batista, P.J. and Chang, H.Y. (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell, 152, 1298-1307.
- Wahlestedt, C. (2013) Targeting long non-coding RNA to therapeutically upregulate gene expression. Nature Reviews. Drug discovery, 12, 433-446.