Esophageal squamous-cell carcinoma (ESCC) is one of the most lethal malignancies in the world and occurs at particularly higher frequency in China. While several genome-wide association studies (GWAS) of germline variants and whole-genome or whole-exome sequencing studies of somatic mutations for ESCC in Chinese population have been published, there is no comprehensive database publically available. Here, as part of collaboration with WU Chen team at the State Key Laboratory of Molecular Oncology, Chinese Academy of Medical Science, we developed the Chinese Cancer Genomic Database-Esophageal Squamous Cell Carcinoma (CCGD-ESCC) database, which contains the associations of 69,593 single nucleotide polymorphisms (SNPs) with ESCC risk in 2022 cases and 2039 controls, survival time of 1006 ESCC patients (survival GWAS) and gene expression (expression quantitative trait loci, eQTL) in 94 ESCC patients. Moreover, this database also provides the associations between 8833 somatic mutations and survival time in 675 ESCC patients. Our user-friendly database is a resource useful for biologists and oncologists not only in identifying the associations of genetic variants or somatic mutations with the development and progression of ESCC but also in studying the underlying mechanisms for tumorigenesis of the cancer.
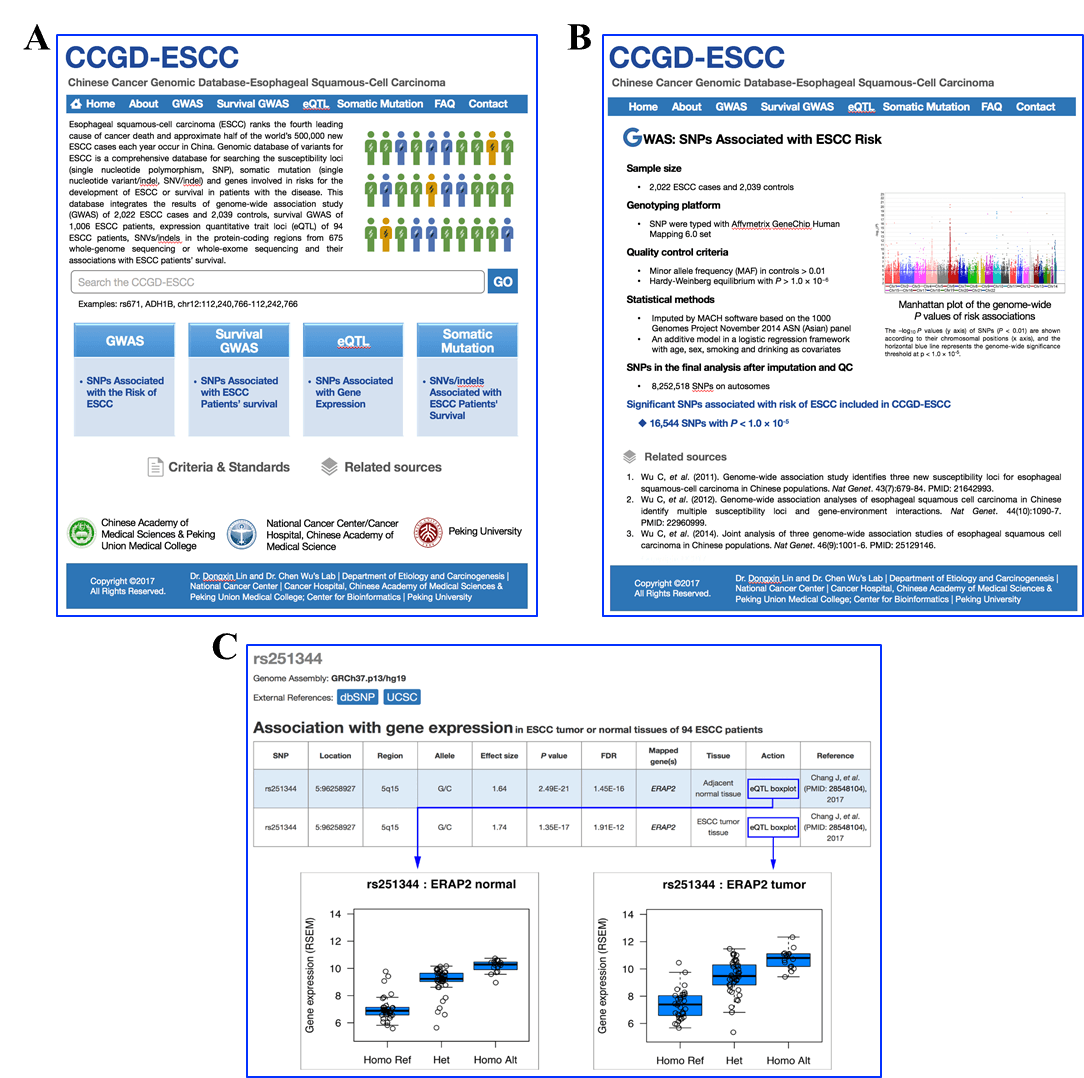
A.Home page. B. Recourse description. C. Query result example.
The paper entitled “CCGD-ESCC: A Comprehensive Database for Genetic Variants Associated with Esophageal Squamous Cell Carcinoma in Chinese Population” is online at Genomics, Proteomics & Bioinformatics (GPB), with the Web server available publically at http://ESCCdb.gao-lab.org
Website: http://ESCCdb.gao-lab.org
Paper link: https://doi.org/10.1016/j.gpb.2018.03.005
Reference:
Wu C, et al. (2011). Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet. 43(7):679-84. PMID: 21642993.
Wu C, et al. (2012). Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet. 44(10):1090-7. PMID: 22960999.
Wu C, et al. (2013). Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nat Genet. 45(6):632-8. PMID: 23644492.
Wu C, et al. (2014). Joint analysis of three genome-wide association studies of esophageal squamous cell carcinoma in Chinese populations. Nat Genet. 46(9):1001-6. PMID: 25129146.
Chang J, et al. (2017). Genomic analysis of esophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat Commun. 8, 15290. PMID: 28548104.
Cancer Genome Atlas Research. (2017). Integrated genomic characterization of oesophageal carcinoma. Nature. 541(7636):169-175. PMID: 28052061.
Qin HD, et al. (2016). Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am J Hum Genet. 98(4):709-27. PMID: 27058444.
Sawada G, et al. (2016). Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology. 150(5):1171-82. PMID: 26873401.
Zhang L, et al. (2015). Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 96(4):597-611. PMID: 25839328.
Gao YB, et al. (2014). Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 46(10):1097-102. PMID: 25151357.
Song Y, et al. (2014). Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 509(7498):91-5. PMID: 24670651.